Scientists at Harvard have successfully encoded an entire digital HTML book including text, images and JavaScript into DNA by using next-generation sequencing technology and a novel strategy to encode 1,000 times the largest data size previously achieved. Deoxyribonucleic Acid or DNA is a nucleic acid containing the genetic instructions used in the development and functioning of all known living organisms (with the exception of RNA viruses). So it can store vast amount of information utilizing only a tiny bits of space and DNA are known to be long lasting, recoverable even after thousands of years.
This was made possible by George Church, the Robert Winthrop Professor of Genetics at Harvard Medical School, and his team who encoded Church’s new book Regenesis: How Synthetic Biology Will Reinvent Nature and Ourselves into DNA.
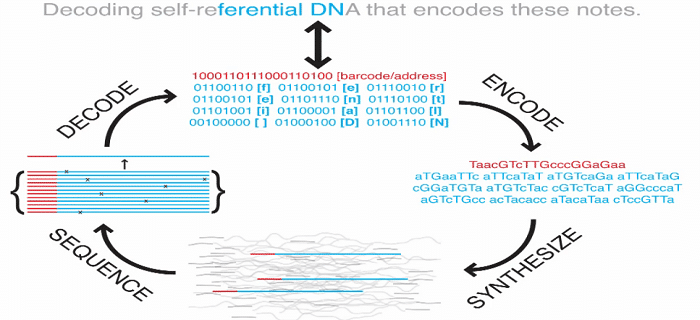
The picture below demonstrates how a text from the book is encoded into binary and then into DNA code.
Commercial DNA chips were used in the process as using live organism would waste much more space because DNA constitute a very small space in a cell and if the DNA doesn’t provide any advantage to the organism, it might completely be deleted by the cell.
Biology’s databank, DNA has long tantalized researchers with its potential as a storage medium: fantastically dense, stable, energy efficient and proven to work over a timespan of some 3.5 billion years. While not the first project to demonstrate the potential of DNA storage, Church’s team married next-generation sequencing technology with a novel strategy to encode 1,000 times the largest amount of data previously stored in DNA.The team reports its results in the Aug. 17 issue of the journal Science.The researchers used binary code to preserve the text, images and formatting of the book. While the scale is roughly what a 5 ¼-inch floppy disk once held, the density of the bits is nearly off the charts: 5.5 petabits, or 1 million gigabits, per cubic millimeter. “The information density and scale compare favorably with other experimental storage methods from biology and physics,” said Sri Kosuri, a senior scientist at the Wyss Institute and senior author on the paper. The team also included Yuan Gao, a former Wyss postdoc who is now an associate professor of biomedical engineering at Johns Hopkins University.
[vimeo 47615970]
What do you think?